Researcher TestimonialsMaddy Parsons, Ph.D.
Group Leader (Royal Society University Research Fellow), Randall Division of Cell and Molecular Biophysics, King’s College London
Systems:
A1Rsi spectral confocal system and Ti-E inverted research microscope with OKOLab environmental chamber and 405, 440, 488, Multiline Argon, 561 and 640 lasers
A1R confocal system with TIRF attachment, Andor Ixon DU897 EMCCD camera, Ti-E inverted research microscope with OKOLab environmental chamber and 405, 440, 488, Multiline Argon, 561 and 640 lasers
1. Please tell us about your research?
We cover two main research fields; the first field focuses on adhesion receptors, such as integrins, and their roles in adhesion to the extracellular matrix, cell signaling and migration. We work with fibroblasts as a normal cell model, and also with different lines of breast carcinoma cells as a model for cancer cells. These different cell lines are known to show different levels of metastatic behavior. In cancer cells, we believe that the amount of time the receptors are in contact with the matrix, and the molecular composition of these adhesions is related to how likely the cell is to invade. We hypothesize that increased stability of adhesions results in a decreased tendency for cells to invade into the matrix. We are researching the molecules that regulate this process with an overall aim of finding novel ways to inhibit cell metastasis.
The second research field uses skin and lung epithelial cells as a model. These cells are generally non-migratory during normal homeostasis on humans, but undergo migration upon insult such as inflammation or wounding. With these cells our focus is studying how they adhere to other cells and to the extracellular matrix, and how this is controlled by external factors. This research helps us to understand wound healing, blistering skin diseases and inflammation in lung disease.
In all of our experiments we try to mimic the in vivo environment using in vitro model systems. The cells are put into a 3D matrix, and we use different models to study different aspects of adhesion. For example, if we were looking at cancer cell invasion out of the vasculature into the extracellular matrix, we would plate cancer cells in a ‘co-culture’ on top of human endothelial cells on a 3D collagen gel to analyze invasion through this ‘barrier’. We can also change the concentration of collagen in the matrix to simulate the different mechanical stresses on the cells that are seen in vivo. In the case of epithelial cells, we can instead plate them on top of a collagen gel containing fibroblasts to mimic the matrix environment within the dermis. These kinds of experiments allow us to study overall effects such as migration, but also smaller details of live individual adhesions all in the same model system.
Historically, experiments in this field were focused around plating cells on 2D surfaces, and there was a strong emphasis on extracting these cells and analyzing them using various biochemical methods. When these cells are in a 3D matrix and in co-culture, it makes it very difficult to extract them, so imaging becomes the method of choice as it gives a better idea of the in situ behavior of the cells. It also allows the capture of dynamics—something not possible with the traditional biochemical approach.
2D microscopy was also used historically to image bulk populations. The ability to image in 3D and 4D allows us to capture information about the z-dimension, as well as capture dynamic events. As these occur over second timescales it is necessary to be able to image at high-speed.
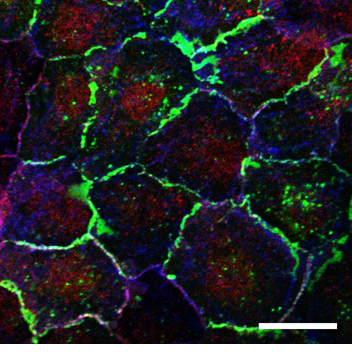
Human lung epithelial cells grown as a confluent monolayer, fixed and stained for e-cadherin (green), CAR (red) and actin (blue). Scale bar 30 µm.
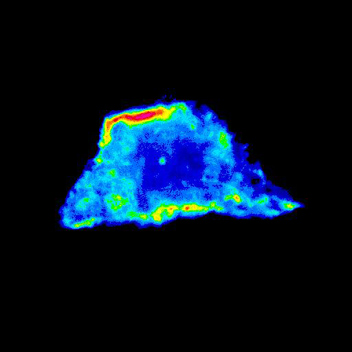
Analysis of RhoA GTPase activation in human epithelial cells using FRET biosensor. Image shows pseudocolor lookup table for RhoA FRET biosensor measured by acceptor photobleaching. Red shows high RhoA activity (20% FRET efficiency) and blue areas are low activity. Note high activity at cell-cell adhesion areas.
2. What applications do you use the Nikon A1Rsi and A1R-TIRF systems for?

The resonant scanner of the A1R allows imaging of short-term dynamic events in live cells over long periods, and allows fast z-stack acquisition. We use the TIRF system for studying adhesion at the coverslip, and the turnover of the adhesion receptor components. We also combine this with XYZ imaging on the A1R in a single experiment so we can follow the fate of the components as they enter the cell.
We use the spectral detector on the A1Rsi for colocalization studies. We could potentially express three fluorophores in the same cell and want to know how these colocalize, especially during dynamic processes. The ability to spectrally image and unmix these fluorophores and remove crosstalk allows us to be confident of our colocalization results.
An example of an experiment unique to the A1R would be the experiments studying dynamic membrane protrusion events. Cells expressing fluorescent probes to label the F-actin cytoskeleton are put in a 3D collagen matrix. These cells form sub-micron protrusions that form and collapse in less than 5-10 seconds, and we believe these dynamic events contribute to cell invasion. Imaging these protrusions requires the acquisition of rapid z-stacks with the resonant scanner and piezo stage. The protrusion dynamics (length, lifetime, etc.) of cells with and without certain proteins are studied, as well as gathering information about invasion of the cell into the matrix over a longer time period to allow us to make correlations between protrusion dynamics and the ability of the cell to invade.
To study proteins that localize to cell contacts and study the recruitment and dynamics of the cell junction we also routinely carry out FRAP analysis of adhesion receptor turnover in epithelial cells. These cell junctions are often destabilized in inflammatory lung disease and the subsequent ‘leakiness’ is associated with an increased inflammatory response. Again, these are highly dynamic, localized processes and therefore require rapid, sensitive imaging approaches to permit accurate quantification.
We also use FRET biosensors to measure activation of proteins in protrusions, for example RhoA GTPase, which controls cytoskeletal dynamics. We look at this activity in various knockdown cells. This is another type of experiment that was historically measured via biochemical methods, but using imaging allows us to measure the localized activation dynamics in much more detail.
3. What was the deciding factor for choosing this Nikon system?
One of the major factors that lead us to choose the A1R was the potential for flexibility in the system; the ability to cover all the types of experiments we wanted in a single microscope, and the possibility to add other components in the future as our research advances. Although there are other microscopes that could do parts of what we required, the A1R allows us to do everything in one system.
4. What was the performance specification that you valued most in selecting this system?
The speed of the resonant scanner was a key consideration for us, as it allows us to capture dynamic events. The ability to flip between the resonant and non-resonant galvano scanners was one of the strongest points, as there is nothing else like it on the market.
5. What do you value about this Nikon system after having used it for three years?
The Nikon Ti Perfect Focus System is excellent and when it comes to timelapse experiments it has changed our lives. The instrument does exactly what we wanted and has significantly increased the research possibilities available to us. We also value very highly the communication with Nikon. Their feedback and enthusiasm for science is something we have not experienced with other companies.
6. How has this system helped your research?
With the A1R systems we have been able to obtain new data that could not have been acquired or analyzed before, and this has led to new and surprising findings. Lots of assumptions have been made in the past from fixed cell studies that this work is now challenging. This has helped re-enforce our position as one of the foremost labs in the world in this area of study.

Human breast carcinoma cell expressing GFP-lifeact to label F-actin, plated on a thin layer of 2D collagen I labeled with Cy3 dye immobilized on a glass coverslip.

Human breast carcinoma cell expressing GFP-lifeact to label F-actin cytoskeleton, embedded in a Cy3-labeled collagen I gel. Images shown are top down projection (main image), or X/Z or Y/Z axis projections through the cell. Note the dramatic difference in morphology of this cell compared to the same cell plated on equivalent 2D surface in the image on the left.
